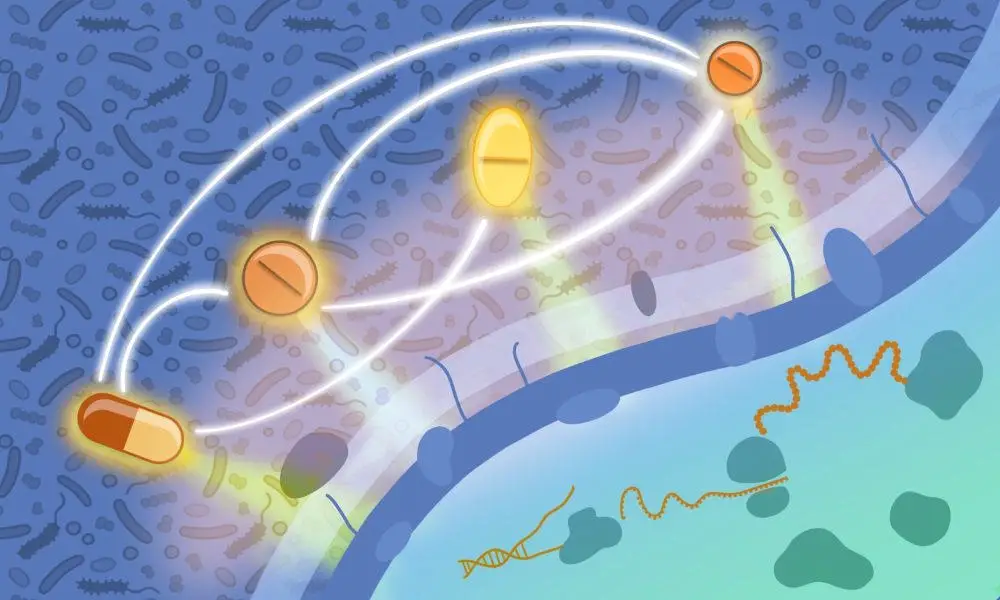
A scientific depiction reveals that antimicrobial medications, which focus on different aspects of bacterial cells, can mutually affect their efficacy. Credit goes to Isabel Romero Calvo and Elisabetta Cacace from EMBL.
In a comprehensive research endeavor, scientists from EMBL have examined over 10,000 pharmaceutical combinations against key pathogenic bacteria that exhibit antimicrobial resistance and are implicated in human mortality.
Antimicrobial resistance, the phenomenon where infectious agents become resilient to antibiotic treatments, remains a significant and urgent global health concern. Research from 2022 demonstrated that nearly five million deaths in 2019 could be attributed to bacteria that are resistant to antibiotics; more than one million of these deaths were directly due to antimicrobial resistance.
In a recent research effort, the Typas Group at EMBL Heidelberg has methodically assessed the effectiveness of over 10,000 pharmaceutical pairings against prevalent multidrug-resistant bacteria.
Elisabetta Cacace, the inaugural author of the study and a former doctoral student in the Typas Group, noted, “While prior studies have explored specific drug pairings, especially those frequently co-prescribed in medical settings, a systematic understanding of how different classes of antibiotics, or mixtures of antibiotics and non-antibiotic drugs, impact bacterial physiology has been lacking.”
Now a postdoctoral researcher at ETH Zürich, Cacace, who also holds a medical degree, has been deeply involved in studying antimicrobial resistance throughout her career. While part of the Typas Group, which focuses on high-throughput techniques for examining bacterial interactions and physiology, her research pivoted towards understanding the mutual effects of antibiotics on their respective cellular targets.
Different antibiotics interact with varying cellular structures or processes within bacteria. These drugs can either synergize, enhancing their collective potency beyond individual effects, or antagonize, where one drug inhibits the efficacy of another. Such antagonistic interactions can be strategically used to minimize the unintended impact of antibiotics on beneficial gut microbes.
In a preceding study, the Typas Group had examined drug combinations against Gram-negative bacteria, which include dangerous antimicrobial-resistant pathogens like E. coli, Salmonella enterica, and Pseudomonas aeruginosa. However, many fatal antimicrobial-resistant bacteria are also Gram-positive, such as methicillin-resistant Staphylococcus aureus (MRSA), which leads to hundreds of thousands of deaths annually. These bacteria differ in cell wall structure from Gram-negative bacteria, affecting drug activity and efficacy.
For the present research, sophisticated robotic setups were used to simultaneously evaluate hundreds of antibiotic and non-antibiotic drug combinations across varying dosages on three representative Gram-positive bacterial species—Bacillus subtilis, Staphylococcus aureus, and Streptococcus pneumoniae. The study included over 8,000 combinations of 65 distinct antibiotics from all major classes, as well as over 2,500 pairings of antibiotic and non-antibiotic drugs, given the increasing prevalence of polypharmacy—the concurrent usage of multiple medications.
By employing this method, the researchers identified over a thousand synergistic and antagonistic interactions, the effects of which were highly species-specific and even strain-specific. These findings diverged from those observed in their previous study on Gram-negative bacteria. Some results were also validated in vivo by infecting moth larvae with pathogens and assessing the efficacy of specific drug combinations in aiding recovery.
The complete database of interactions has been made publicly accessible for scientists to review, investigate, and search for new synergistic or antagonistic interactions.
“The sheer scale of this study makes it exceptional. The data set is so extensive that it will likely provide fodder for hypothesis generation for years,” remarked Cacace. “The study is also enlightening from a systems biology standpoint, unveiling drug interactions targeting cellular processes previously unknown.”
Nassos Typas, the senior author of the study and EMBL Group Leader, concluded, “We are in a period where innovative approaches to counter antimicrobial resistance are critically needed. The development of new antibiotics is both technically complex and resource-intensive. The systematic profiling of drug interactions we’ve conducted offers an avenue for alternative treatment strategies for bacterial infections.”
Reference: “Systematic analysis of drug combinations against Gram-positive bacteria” by Elisabetta Cacace et al., published on 28 September 2023 in Nature Microbiology.
DOI: 10.1038/s41564-023-01486-9
Table of Contents
Frequently Asked Questions (FAQs) about Antimicrobial Resistance
What is the main focus of the research conducted by EMBL scientists?
The main focus of the research is to evaluate the effectiveness of over 10,000 drug combinations against pathogenic bacteria that exhibit antimicrobial resistance. The study aims to understand both synergistic and antagonistic interactions between different classes of antibiotics and between antibiotics and non-antibiotic drugs.
Who conducted the study and where was it conducted?
The study was conducted by researchers from the Typas Group at EMBL Heidelberg. Elisabetta Cacace, a former doctoral student in the Typas Group and now a postdoctoral researcher at ETH Zürich, was the inaugural author of the study.
Why is this research important?
The research is crucial because antimicrobial resistance is a significant global health issue. It threatens the effectiveness of existing antibiotics and poses a severe risk of increased mortality from bacterial infections. This study provides systematic insights into alternative treatment strategies.
How many drug combinations were tested in the study?
Over 10,000 drug combinations were tested, including more than 8,000 combinations of 65 different antibiotics spread across all major classes, and over 2,500 combinations of antibiotic drugs with non-antibiotic drugs.
What types of bacteria were targeted?
The study targeted three representative species of Gram-positive bacteria: Bacillus subtilis, Staphylococcus aureus, and Streptococcus pneumoniae. It extends previous research that focused on Gram-negative bacteria.
What were the significant findings of the study?
The study identified over a thousand synergistic and antagonistic interactions between drugs. These interactions were highly species-specific and even strain-specific. The findings could pave the way for alternative treatment strategies against bacterial infections.
Is the database of drug interactions publicly accessible?
Yes, the complete database of interactions has been made publicly accessible for scientists to review, investigate, and search for new synergistic or antagonistic interactions.
What are the future implications of this research?
The extensive dataset generated from this study will likely serve as a basis for further research and hypothesis generation for years to come. It opens up the path to alternative solutions and treatments for bacterial infections, particularly in the face of increasing antimicrobial resistance.
Who are the intended beneficiaries of this research?
The intended beneficiaries are the global healthcare community, pharmaceutical researchers, and ultimately patients suffering from bacterial infections that have become resistant to existing antibiotic treatments.
What makes this study distinct from previous research?
The sheer scale of this study sets it apart, and it systematically profiles drug combinations in a way that has not been previously conducted. It also expands the focus from Gram-negative to Gram-positive bacteria, thereby broadening the understanding of antimicrobial resistance.
More about Antimicrobial Resistance
- EMBL Heidelberg
- Typas Group Research
- Antimicrobial Resistance Global Report
- Systematic Analysis of Drug Combinations Against Gram-Positive Bacteria – Nature Microbiology
- Polypharmacy in Modern Medicine
- ETH Zürich Research
- Global Health Challenges
- Staphylococcus aureus Infections
- Systems Biology Research
- Gram-Negative vs. Gram-Positive Bacteria


5 comments
Its just scary to think almost 5 million people died due to antibiotic resistant bacteria in just one year. Research like this is not just valuable, it’s vital.
Wow, this is groundbreaking stuff! The scale of the study is just immense. Over 10k drug combos? That’s next level. Antibiotic resistance has been a ticking time bomb, and this might be a key to defusing it.
Gotta say, the data being publicly available is awesome. More brains on this problem the better. Open science ftw.
incredible work by the EMBL team. Couldn’t be more timely with antibiotic resistance on the rise globally. Our existing tools are blunt, we need more precise ways to fight bacteria.
What struck me was that they even considered non-antibiotic drugs in the combinations. Shows they’re really thinking outside the box. Could be a game changer in an age where people are on multiple meds.